Introduction
The idea behind ex-novo assisted 3D elucidation is to process a set of spectroscopic data obtained from a sample in order to describe all the possible molecular structures consistent with it while requiring a minimum amount of human intervention.
Our approach in this introductory article will deliberately be focused on giving an overview of the determination of stereochemistry/conformation of completely unknown compounds. These may have been isolated from natural products, or novel, synthesized molecules. As such, the process falls short of full 3D structure elucidation, but is an important point on the road to this larger objective.
Further articles will describe and illustrate how the intermediate challenges are dealt with, to explain how the whole block diagram gets defined, and how the component parts fit together. Significant advances have been made in this field in the past few years, and software has been developed to address more or less successfully many of the associated intermediate goals. Nowadays it is easy to find in the web a number of comprehensive reviews on the subject (1) (2).
Setting the scene
The following terms describe the more common and relevant procedures associated with the understanding and use of spectroscopic data and their associated models:
- Structure characterisation, relates to collecting spectroscopic data (IR, UV, NMR, MS, X-ray) related to a specific molecular structure. It does not necessarily involve complete interpretation of these data.
- Structure verification (ASV) and structure integrity confirmation (ASIC), involve using some or all of these data to confirm a proposed chemical entity or group of chemical entities. The result may be as simple as confirmed or not confirmed.
- Structure elucidation, relates to the determination of molecular structure(s) without preconception through something like an assisted synthesis process. Entire structures or fragments thereof may be determined on the basis that the experimental data are consistent with the theoretical description of derived data for the arrangement of atoms in that part of the molecule. A full theoretical analysis may not always be possible, and the door is opened to the possibility of assisted expert (human) knowledge input. No consideration is given for whether the atom arrangements in the fragments are energetically feasible.
What should we be looking for?
Put simply, we are aiming for data sets encoded with the spatial coordinates of the essential molecular components (atoms) and their bonding information. But achieving this is far from trivial!
Instead of thinking of this endeavour as a methodical search for certainties, it could rather be regarded as an iterative process to add and verify positive hypotheses, based on a series of automatically extracted and user-contributed clues that converge upon a solution. The completeness, or success of this process may depend on the available – possible – data that can be derived for the particular chemical system.
Plan sketch
We shall start with preprocessed spectroscopic data and then describe or extract the relevant experimental signals. Then we will associate these with the established molecular model, allowing for contributions from user experience and/or insights. The last step will comprise building a rough structure (with one or many fragments), and arranging and shaping them in such a way that a merit function is optimised. The merit function initially will sum up contributions derived from the interatomic distances (accurate or not) that could be deduced.
To be precise in setting the scene, let us make clear that no fragment-based screening based on chemical shift prediction is intended in this plan, even in the case of underdefined problems (those with very few protons). It is important to be explicit on this as, logically, a combination of 3D and fragment-based methods will be the way to solve the widest number of problems.
Interatomic distances
Structure can be sufficiently described by a minimum set of interatomic spatial distances. These are themselves dependent on chemical bond lengths, bond angles (between 3 atoms), and torsions (angles plus dihedral angles, between 4 atoms):
- Chemical bond lengths depend on the atom radius and the type of orbitals involved - they vary slightly in time (5% at room temperature) due to thermal oscillations.
- Bond angles are determined and/or influenced by hybridization of the intervening atom orbitals, electron cloud effects and inter-atomic repulsions.
- Dihedral (torsion) angles, unlike bond angles (with only local influence), show their influence over larger distances and define the whole molecule conformation.
Our first task will be to obtain a set of (inaccurate) interatomic distances with these properties:
- Each distance will comprise a value and a type:
- The value will be taken from tables (e.g., bond distances from X-ray crystallography) or calculated from the spectroscopic data (e.g., interproton distances derived from nuclear Overhauser effect (NOE) experimental spectra).
- The type will be assigned (from a set of predefined values) according to the modelled interaction or interactions corresponding to the nature of each experiment's signals.
- Each distance value will contribute to the merit funcion in a predefined way according to its type.
Available spectra and required spectra
The information we want to extract is encoded in various kinds of spectral information recorded in different experiments, normally done with the same (chemically and physically stable) sample.
The state-of-the-art in NMR presents us with a wealth of experiments and sophisticated instrumentation. We shall shortly be describing what comprises a minimum set of spectra amenable to the start of an ex-novo elucidation.
Let us first take a look at the characteristics of the most important NMR experiments in this context (this is a less abridged account and some updated guides are avaliable in the web (3)):
| Experiment | Information | Derive |
| 1D proton (1H NMR) | Chemical shifts of proton resonances. Relative intensity (area) of peak/s. Splitting pattern due to scalar spin-spin couplings between protons typically separated 2-4 bonds. | Chemically distinct loci within compound, corresponding chemical shift values, and relative nbr. of proton(s) in each of them. |
| 2D 1H-1H COSY - Homonuclear Correlated Spectroscopy (and variants) | Nuclei sharing a scalar (J) coupling (usually hydrogen, but could be any high-abundance homonuclear spins like 19F, 31P ) | Vicinal Hs (or other). C-C bonds, by inference. |
| 2D 1H-1H NOESY and ROESY - Nuclear Overhauser Enhancement Spectroscopy | Proton-proton correlation (mediated by dipolar, through-space coupling) | Interproton distance |
| 2D X-1H H2BC - Heteronuclear 2-bond Correlation | Medium-range correlation of 1H and a heteronucleus | Vicinal Hs and C-C (or corresp. heteronuclear) bonds. |
| 2D X-1H HSQC - Heteronuclear Single Quantum Coherence | Single quantum coherence between J-coupled protons and heteronuclei:- proton magnetization (transverse) set to zero during evolution phase- X nuclei BB-decoupled during proton acquisition | Hydrogen and heteronuclei one bond apart. C-H bonds. |
| 2D X-1H HMBC - Heteronuclear Multiple Bond Correlation | Zero and double quantum coherence between J-coupled protons and heteronuclei | n-bonds apart (2 - 4 or more) hydrogen and heteronuclei |
Dissecting our plan: Acquire - Analyse - Use model - Build structure.
1 - Acquire: Acquire spectroscopic data from relevant spectra
The bare minimum set of spectra that makes 3D elucidation feasible should provide:
- A way to enumerate or assign (we shall use both terms interchangeably) as many NMR active nuclei in the compund as possible: some hydrogens and some heteroatoms (i.e. other nuclei than H) should be enumerated.
- A way to establish some relations or connectivities between them, translatable into interatomic distances: Some connectivities relating heteroatoms (C, N, ...) with Hs should be proposed.
A first approach could begin by acquiring a 2D HSQC X-1H spectrum, which ideally would show all signals corresponding to X atoms (usually C or N) directly bonded to one or more Hs. This would effectively serve to relate (and impose distances between) heteroatoms and Hs. However,
- this (alone) would not show any clue to locating heteroatoms with respect to each other, and therefore would not be conducive to obtaining even a very coarse, overall scaffold of a molecule or of any fragment thereof, and
- the risk would exist that some spurious or systematic artefacts would inadvertently mislead us, with no chance to do any cross-checking with other spectra.
We should therefore obtain at least:
- a 2D HSQC, plus a 1D proton spectrum. The latter adds basic but important information about protons, and allows a GSD-based automatic multiplet analysis
- a C-C (or N-N) revealing spectrum like 2D COSY or H2BC
- at least one other experiment like 2D HMBC or 2D NOESY/ROESY that provides information over medium distances.
- Deciding which other, more complete sets of spectra to acquire very much depends on each particular case - the known compound characteristics to be discovered, and the analysis efforts already done!
- For example, if one can at all expect nitrogen to be a key element in the compound backbone, then a 15N-1H HMBC and/or HSQC spectrum would be an obvious choice.
- However, if one suspects that the molecule consist of a number of separate spin systems, then 2D ROESY or NOESY could provide clues to link them, or at least place them with respect to each other.
- Another axample is the search for the structure of small molecules having a relatively high proton density, and for which resonance overlap is expected: in such cases homonuclear correlation experiments like 2D COSY or TOCSY would surely be valuable.
2 - Analyse: Select relevant signals and extract relevant information.
Referencing spectra
Referencing (or aligning) spectra may seem unimportant or trivial to mention, but because it is absolutely critical for 3D elucidation we feel we must comment on it.
It is very important to consider that in the realm of ex-novo elucidation the goal is not probing the expected existence or absence of a signal in a spectrum in order to confirm or disprove something. Rather it is about being faced with an experimental signal of unknown origin, and bearing in mind that that signal will mean something valuable inasmuch as it is either clear-cut and/or also clearly identified in other spectra, whether directly or on the basis of deductions from the model.
Referencing is so important that the very process of assisted or incremental peak picking, (see below), has an implicit and very convenient method to account for the very small misalignment that may happen in the values and/or the graphical representation of signals after the acquisition and preprocessing stages. In fact, this is not uncommon.
The user must therefore ensure that all the spectra are properly referenced with respect to a reliable reference spectrum (Mnova provides an 'Absolute Reference' procedure for referencing all spectra in the document by using the 1D proton spectrum (if present). This can be used to reference all 1H and X-nucleus spectra in 1D and 2D cases. The exact referencing problem is exacerbated in 2D spectra, where digital resolution is often coarse in one dimension, but can in some cases be effectively mitigated by using processing methods such as zero filling or linear prediction - the outcome of these improvements then prompting for a refined referencing.
Selecting signals - Peak-Picking, or the critical link between measurements and model
This is a huge subject itself and this section intends just to touch on the main ideas.
The goal of peak-picking is to distinguish signals and afford chemical shift information of the interesting spin systems in the noisy NMR spectra, while ignoring irrelevant or confusing signals from impurities, artefacts, solvent signals, etc.
It is a mandatory and sensitive stage, and must be done very accurately, as it defines the atom(s)-to-chemical-shift assignments and the relations or connectivities between them.
- The first 2D spectrum to be peak picked should be 2D HSQC, and then the derived chemical shift assignments can be superimposed over the other spectra when doing the corresponding peak picking (i.e. either manually, or when reviewing one that was peak-picked automatically). At this point the user will decide, many times taking into account the aforementioned misalignments, if a signal is bona fide and should be picked or kept.
- Of course peak picking must be done bearing in mind the nature of the spectrum being treated. For example, HMBC spectra accounts for quaternary carbons, but these are not visible in HSQC, so one could pick an HMBC peak while not having a nearby HSQC carbon assignment. Just to make the most of this example, there is a good likelyhood that some carbon satellite signals appear clear and strong in HMBC, but in this case these should not be peak picked, or better yet, picked and tagged as artefacts. These can usually be recognised, as they must be related to a carbon assignment and be symmetrically placed about the single-quantum signal.
The main difficulties are:
- Well-defined signals are strong (good s/n) and clear-cut, but some are not. So - sometimes it can be difficult to set the thresholds. Connectivity building will be hampered if wrong assertions are made by picking wrong signals.
- The ability to attain a realistic outcome will be severely hampered if the user and/or the software is not good enough at discerning signals pertaining to the physical system under study from the background or spurious ones.
3 - Use model and user's experience and insight.
Atom enumeration
After finishing the peak-picking stage we can produce a list of some of the components of the molecule of interest. Atom enumeration is about assigning labels to the different chemical shifts found for the NMR active nuclei detected and recorded in the spectra.
An NMR spectrometer records signals produced collectively from an ensemble of physical nuclei in a sample. This sample must be prepared such that a sufficient concentration of the interesting compound/s is present and is chemically stable through the course of the measurements. Usually the compound is chemically pure to a level in excess of ca. 95%, and a fully deuterated solvent is used.
Under the assumption that the outcome of the sample preparation work is a stable, limited number of different compounds present at high concentration in the sample, one can expect that the detected, measurable chemical shifts correspond to (NMR active) nuclei present in these compounds.
So the assertion can be made that the minimum number of nuclei in a chemical species is equal to the number of observable multiplets. A more complete picture emerges when multiplet integrals are considered.
Connectivities
Connectivities are just relations between 'enumerated' atoms, established on the basis of the signals picked in spectra, or assignments made by the user. They are grouped in a connectivity table (CT)
We must also distinguish two kinds of connectivities that comprise a standard CT:
- 'primary' connectivities. These are the ones derived directly from the user-peak-picked spectra, or deduced from them by a set of rules reflecting current knowledge
- 'secondary' connectivities carry less weight. They may be user-introduced connectivities or derived automatically from other secondary using rules.
Secondary connectivities are intended to let the user introduce relations that cannot be reported in the available spectra. The user might know that a non-detectable relationship exists, and decide to make the corresponding assertion by introducing a secondary connectivity. Or the user could be interested in testing some tentative relation: - for example, to join molecule fragments, he/she would introduce some secondary connectivities, and then carry on with the elucidation to see if the outcome is sensible and in accord with the data.
Rules and filters
Let us remind ourselves here that a set of interatomic distances (value and type) must exist before structure generation can occur.
To achieve that, we must ensure that most of the knowledge about molecular structure is included in our model, and deduce as many connectivities between pairs of atoms as we can. The strategy is based on taking an (automated) look at the current connectivities and applying a set of filters and consistency checks upon them, and rules to deduce new ones.
Let us illustrate this idea with some examples. The following illustration (Fig.1) shows the rule to deduce a primary C-C connectivity (strongly resembling a C-C single bond) and having a defined (tight) interatomic distance:
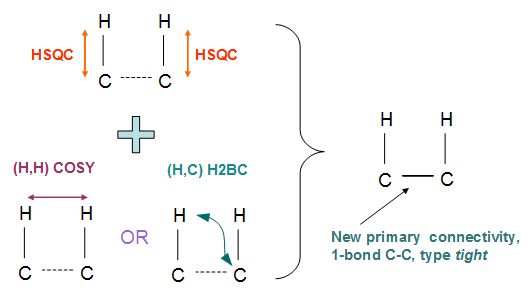
Fig. 1. Schematic of a rule to obtain a new primary connectivity from two existing ones. In this case they come from combining either HSQC+COSY or HSQC+H2BC observations.
In Fig. 2 we show the rule to deduce two primary connectivities - one of type tight and other say semi-loose - from two existing ones:
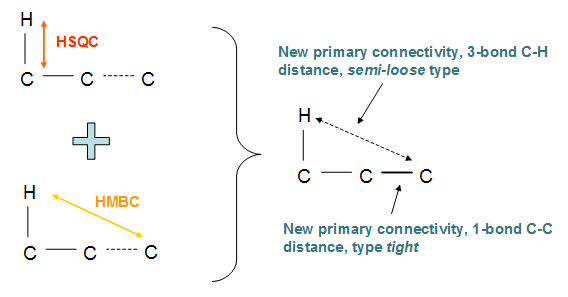
Fig. 2. Rule to obtain two new (primary) connectivities from two (primary) derived from 2D HSQC and HMBC data.
We could also consider how secondary connectivities may be introduced by the user. For example, if an (H,H) TOCSY spectrum is available but its resolution only scarcely allows to identify some separate spin systems with high uncertainty, then the user could decide to try introducing certain secondary C-C connectivities (i.e. 1-bond C-C) between the corresponding protonated carbons of the possible spin systems and see if the outcome makes sense. And this is indeed a common example, as the homonuclear (H,H) COSY spectrum often has insufficient resolution to allow a reliable peak-picking and assignments to be performed when many proton resonate close to each other!
- It is important to mention here that rules and filters must apply to both primary and secondary connectivities. That allows one to quickly and productively test the outcome of the user's insights in enriching and/or complementing the information that can be extracted from spectra through seemingly relevant peak-picked signals.
4 - Build structure (or conformation).
Again, structure generation is a huge subject on its own. An intentionally very brief overview is as follows:
- Once a set of distances is calculated, the task is to find the global minimum of a real-valued, nonperiodic, merit function [hyper]surface that is dependent on them and of course the spatial coordinates of the components (nuclei). This uses the Distance Geometry algorithm.
- The merit function comprises a set of sub-functions which weight the contributing values of the various types of distances. Distances may be quite crudely categorised based on whether they are “tight-valued”, “flexible-valued”, or intramolecular "long-range".
- Many intermediate local minima basins occur in the surface, and the algorithms must 'hop' between many of these and find a number of low-valued outcomes - and eventually a global minimum.
The Distance-Geometry Module
Distance Geometry or DG involves calculations where consideration for the spatial coordinates alone is given for the atoms. How these atoms may be connected with bonds is not considered. In the context of NMR and this exercise, the most relevant input data are the internuclear separations and how these may be adjusted so they are consistent with known bond distances and those derived from nuclear Overhauser effect (NOE) measurements.
Some analytical expressions account within DG for the various distance types considered. The distance geometry algorithms try to arrange atoms in 3D space so that the distances derived from experimental data and contributed by the user fit as well as possible with nuclei spatial coordinates.
In conclusion, we have started to lay down the sequence of steps and experiments necessary for this unique approach to structure/conformation elucidation. The practicalities are fraught with difficulties, and we touch on these only briefly for now. But the outcome is satisfyingly (surprisingly?) effective. Successive articles in this series will deal with the steps in more detail and build up a complete picture of the process.
Acknowledgement
The author has benefitted to a large extent from numerous discussions with Drs. Craig Butts and Jeremy Harvey of School of Chemistry - Univ. of Bristol, and with Dr. Manuel Martín of NMR Service - Univ. of Santiago de Compostela. Their collaboration and continuing scientific input is gratefully acknowledged.
- Bross-Walch, N., Kühn, T., Moskau, D. and Zerbe, O. , Strategies and Tools for Structure Determination of Natural Products Using Modern Methods of NMR Spectroscopy. Chemistry & Biodiversity, 2 2005: 147–177. doi: 10.1002/cbdv.200590000 http://onlinelibrary.wiley.com/doi/10.1002/cbdv.200590000/pdf (accessed Jun 11, 2012).
- Mikhail Elyashberg, Kirill Blinov, Sergey Molodtsov, Yegor Smurnyy, Antony J Williams and Tatiana Churanova. Computer-assisted methods for molecular structure elucidation: realizing a spectroscopist's dream. Journal of Cheminformatics 2009, 1:3 doi:10.1186/1758-2946-1-3 http://www.jcheminf.com/content/1/1/3 (accessed Jun 11, 2012).
- University of Oxford. Chemistry Research Laboratory. NMR facility. A chemist's quick guide to NMR acronyms and experiments. http://www.chem.ox.ac.uk/spectroscopy/nmr/acropage.htm (accessed Jun 11, 2012).