Introduction
The assignment of 1H NMR spectra of small molecules is an everyday task within organic chemistry, which is usually tackled in a manual way. The chemist typically identifies the most relevant regions (aka multiplets) in the spectrum and assigns them to atoms in the putative molecular structure. A partial assignment is usually attempted and the process generally lacks rigor. This process is generally considered repetitious, time-consuming, very tedious and error-prone.
 Whilst a variety of computational methods to automatically assign NMR spectra of biomolecules have been in use since the early 90s, approaches for the unattended assignment of 1H NMR spectra of small spectra have been more sparse. Of course, having the ability to fully automatically elucidate an unknown structure from just a 1H NMR spectrum would be the ultimate goal of any computer based expert system. Unfortunately, such a tool is only available in CSI (although it’s not uncommon that scientific progress was first visualized in fiction!), so for now we will have to settle for applications that can be used to assign known molecules to their corresponding 1H NMR. And this is, in our opinion, already a very valuable tool, especially today, when the volume of acquire data has increased inversely to the amount of analytical human resources and (sometimes) training.
Whilst a variety of computational methods to automatically assign NMR spectra of biomolecules have been in use since the early 90s, approaches for the unattended assignment of 1H NMR spectra of small spectra have been more sparse. Of course, having the ability to fully automatically elucidate an unknown structure from just a 1H NMR spectrum would be the ultimate goal of any computer based expert system. Unfortunately, such a tool is only available in CSI (although it’s not uncommon that scientific progress was first visualized in fiction!), so for now we will have to settle for applications that can be used to assign known molecules to their corresponding 1H NMR. And this is, in our opinion, already a very valuable tool, especially today, when the volume of acquire data has increased inversely to the amount of analytical human resources and (sometimes) training.
As a response to this necessity, we have developed an expert system for the automatic assignment of 1H NMR spectra of small molecules. This uses the principles of fuzzy logic and probabilistic methods to first classify all the resonances (peaks) in the spectrum and then proceeds to enumerate the most likely assignments of experimental multiplets to a presumed molecular formula, and finally applies a score to them. It uses as inputs the experimental spectrum (or possibly various kinds of spectra spectra), the suggested molecular structure, and the predicted NMR parameters (shifts and coupling constants) and, as output, it generates the most likely assignment.
How does Auto Assignments work?
The Auto Assignment Algorithm combines several software techniques we had developed in recent years as tools for expert tasks such as automatic detection and characterization of spectral peaks, automatic solvent detection, and automatic structure verification (for which the auto-assignment feature is, in its own term, a building block).
Real-life spectra always contain a number of artifacts such as noise, baseline distortions, relaxation and radiation-damping induced distortions of peak intensities, lineshape distortions due to magnetic field inhomogeneity, lineshape distortions due to unresolved weak long-range couplings, second-order interactions, peaks crowding causing peaks and multiplets to overlap, etc.
For these reasons it is impossible to construct any NMR-data evaluation wizard, like the automatic assignment module, without an extensive usage of statistical methods, allowing for a degree of logical “fuzziness”. In our case this is done by applying at every step, to the full depth of the algorithm, a proprietary scoring system approach. A description of such scoring system is beyond the scope of this document, but it will be covered in a future article.
The Auto Assignment algorithm consists of the following constituent blocks (See Fig. 1)
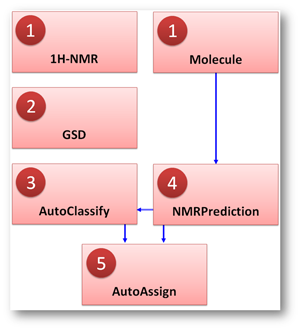
Fig. 1: Basic flowchart diagram of the new 1H-NMR Automatic Assignments algorithm. See the text for a description of its constituent blocks
(1) Basic processing.
An NMR-FID is loaded, apodized, transformed, phased and baseline corrected, typically in a transparent, fully unattended way (The process, however, can be customized by the user).
In addition, a presumed correct molecular structure is loaded, using any of the popular formula-encoding formats (mol, ChemDraw files, etc)
(2) GSD.
The resulting frequency domain 1H spectrum is automatically deconvolved using the sophisticated Global Spectrum Deconvolution algorithm in order to generate a reliable list of peaks and their parameters (position, height, width, kurtosis, area, etc), even in situations characterized by a strong peaks overlap (Fig.2).
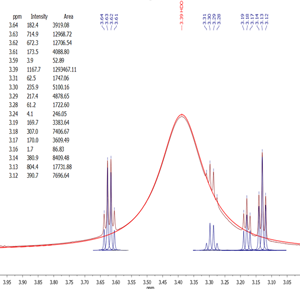
Fig.2 Example of information about the spectral peaks extracted by GSD in the presence of a strong overlap with large, broad water signal.
(3) AutoClassify.
Using another sophisticated fuzzy-logic algorithm, each peak in the GSD list is classified according to whether it belongs to the compound, solvent, an impurity, an artifact, a 13C satellite, etc (Fig.3). The algorithm even attempts to pinpoint possible labile peaks.
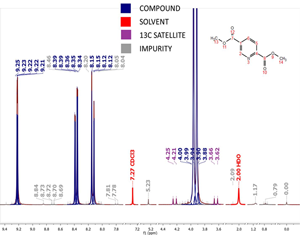
Fig. 3 Illustration of the AutoClassiffy algorithm. Peaks are color coded according to their their type
An important part of this process is also the recognition of multiplets due to J-couplings and a detailed characterization of their many properties (this results in a multiplets list). Inter-multiplet coupling patterns are also detected and stored as another tool for the subsequent auto-assignment step.
(4) NMR Prediction.
NMR spectral parameters (chemical shifts and scalar coupling constants) of the suggested molecule are predicted using three complementary approaches: 3D conformer, substituent chemical shift and a HOSE code database which are then combined using the NMRPredict Best Algorithm which is seamlessly integrated within Mnova software. Users can also add their own assignments to the HOSE code database to further refine the accuracy of the predictions.
(5) AutoAssign.
The final step of the algorithm consists in combining all the information collected to this point. Basically, the wizard tries to find the best possible match between the experimental multiplets and the predicted multiplets, subject also to constraints dictated by NMR know-how. Mathematically, the number of possible assignments is staggering, but we apply a prior enumeration filter passing only a limited number (about 100) of the most likely ones. In this way it becomes feasible to score each assignment against all available information and select the best one.
A more in-depth description of the inner details of the auto assignment algorithm will be presented elsewhere. However, we hope that the present description provides a sufficiently clear picture of its underlying concepts and its most important features.
A simple example
In Figure below, we show the result of applying the new assignment algorithm to the spectrum of L-Proline. The result has been obtained in a fully automatic operational mode; simply drag and drop the molecule and the spectrum and run the command, that is it. Several points are worth nothing:
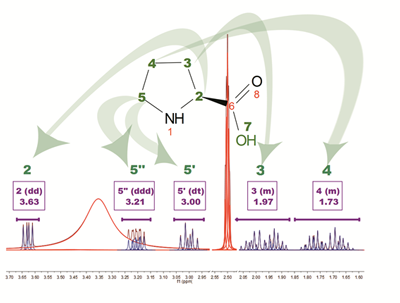
Fig. 4.: Result of running AutoAssignment with L-Proline showing how GSD helps to resolved overlapped issues as well as to classify peaks according to their type. Blue peaks correspond to the detected compound resonances, whereas red lines are signals identified as solvent (DMSO and water).
- Solvent peaks (DMSO and water) have been automatically detected and displayed in the spectrum as red peaks superimposed on top of the experimental peaks.
- Compounds peaks are displayed as blue curves. Notice that, thanks to the power of GSD, it is possible to quantify the peaks corresponding to H-5’’ despite the significant overlap with the large solvent (water) peak. Traditional peak picking routines would fail in cases like this.
Of course, even though this example illustrates some interesting challenges that were successfully overcome, like the ability to yield accurate multiplet integrals even in those cases where extra signals (e.g. solvent) overlap with the peaks or multiplet of interest, by no means this is a system that will yield all the assignments with a 100% success ratio. Certainly, there always will be cases of partial misassignments. In order to assess the number of assignments that a user would typically have to manually amend, we have conducted a test using fully assigned in-house 1H-NMR library consisting of 39 molecules with a total of 355 proton assignments. This test showed that 295 assignments were correctly identified whilst 60 were wrong, corresponding to a 80% success ratio. Very often, the errors were due to two assignments that have to be swapped, a feature that can be done in the software with just two mouse clicks.
Conclusion
In this document we have presented the basic concepts around the new automatic assignment module included in Mnova NMR 8.0. It has been intended mostly as a tool for those organic chemists that have to face routinely to the tedious task of assigning their 1H spectra. The results that we have obtained are, in our opinion, very promising and we believe that it should already constitute a real time saver.
In addition to the automatic facilities provided, the software includes a number of graphical features that facilitates enormously the manual correction of any potential errors made.
In this work, only 1D 1H NMR spectra were used but the system is already armed to accept HSQC spectra. The results obtained with a combined 1H & HSQC approach will be covered in a separate publication.
[1] Gronwald, W.; Kalbitzer, H.R. Automated structure determination of proteins by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2004, 44, 33-96. DOI:10.1016/j.pnmrs.2003.12.002[2] Griffiths, L.; Beeley, H. H.; Horton, R. Towards the automatic analysis of NMR spectra: Part 7. Assignment of 1H by employing both 1H and 1H/13C correlation spectra. Magn. Reson. Chem. 2008, 46, 818-82. DOI: 10.1002/mrc.2257[3] Cobas, c.; Sykora, S.The Bumpy Road towards Automatic Global Spectral Deconvolution (GSD), 50th ENC Conference, Asilomar, CA (USA), March 29-April 4, 2009 DOI: 10.3247/SL3Nmr09.003[4] Kurtosis is one of the pure shape functions that we use to describe a multiplet mathematically[5] Cobas, C.: Seoane, F.; Domínguez, S.; Sykora, S. A new approach to improving automated analysis of proton NMR spectra through Global Spectral Deconvolution (GSD). Spectroscopy Europe, 2010, 23 vol 23 (1), 2010 [Online]