Preceding articles in this series have largely dealt with aspects of data acquisition to ensure reliable quantitation by NMR. A script was described that can be used to determine compound purity by manually selecting integration regions for the concentration reference compound, and the compound of interest. Clearly these are strictly manual procedures are totally subjective. But what if we would like to analyse NMR data to perform qNMR – automatically and objectively?
The move to automated qNMR is desirable for a number of reasons:
- a laboratory may collect high volumes of NMR data and manual analysis would be almost impossible
- quantitation can be performed at the same time that other automated analyses are performed, e.g., Verify
- a reliable algorithm for multiplet selection can outperform relatively novice users and consistently perform very well
- Apply GSD peak picking and peak line classification
- Identify the multiplets in the spectrum and determine their type (doublet, etc.) and the number of nuclides each represents.
- We then calculate AI/NN for each multiplet - the Absolute Integration divided by the number of nuclides it represents.
- A quantitation number (e.g., concentration) is calculated for each multiplet
- The best multiplets are chosen to provide an average result and RMSD%
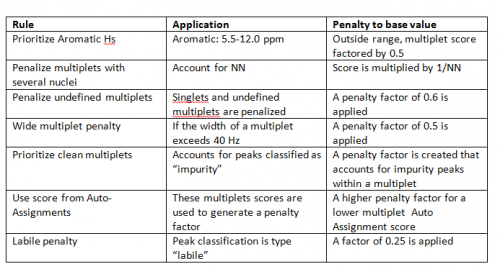
Note that “Auto Assignments” are very effective with steps 1-3. Furthermore, this analysis provides a score for each multiplet that we can use (see below). The score accounts for a number of relevant factors for a multiplet, including penalties for impurity peaks.
Whilst each of these steps is a subject in its own, we will concentrate in this article on the fundamental stage of multiplet selection. This is a key step in a number of related tasks and therefore deserves discussion in its own right.
Choosing the best multiplets
Whilst the reasons behind these options may vary depending on the user’s typical spectra, the following shows how multiplet ranking and minimum RMSD can be used.
Choosing multiplets:
- Multiplet ranking
- Minimum RMSD
- Ranking/elimination followed by Minimum RMSD

Minimum RMSD
This process, very simply, chooses the multiplets that yield an average value having the smallest RMSD% - the RMSD normalised to the mean of the observed values. The user specifies the number of multiplets to be used.
Multiplet ranking
We have devised a scoring system based on a variety of user-selectable criteria. If a criterion is selected and a multiplet violates it then that multiplet’s score is factored down accordingly. For example, peaks from labile protons are almost always poor candidates for qNMR because they frequently under-integrate. If a multiplet is ascribed to a labile proton then a (severe) factor is applied to its base score, making it almost impossible for this multiplet to be selected.
Base score
Each multiplet starts with a score of 1.0, and this value is then modified based on how close its qNMR value is to the average concentration for all multiplets. In this way clear outliers receive an immediate penalty before any of the considerations, below. This is the base value for a multiplet.
Scoring rules
The scoring rules are described in the table, below.
Final multiplet score
Penalties are multiplicative and each is applied to the base score for each multiplet to derive a final score.
Score threshold
The user can specify a final score threshold, and highly penalized multiplets will thereby not be considered for selection.
Hybrid method
This is an amalgamation of multiplet scoring and RMSD minimisation methods. The scoring system and threshold are used to reject poor multiplet targets, and then the minimum RMSD method is applied to the “survivors”.
What, when, and why?
So why would you want to bother with all this? Why not simply set to RMSD minimum, and that’s it?
The answer to these questions really comes down to a number of considerations that may relate specifically to your data, and the process of deselecting multiplets that are contaminated with peaks from impurity materials. Some common problem situations are shown in Figs 1-3, demonstrating the need for a considered and flexible approach to the parameters that should be selected. We see examples that lead the user to selecting appropriate parameters for the general issues seen with typical samples that must be analysed.
Fig 1. An example of a “wide multiplet” (>40 Hz). Could some peaks be from impurities? Here a “Wide multiplet” penalty would be appropriate.
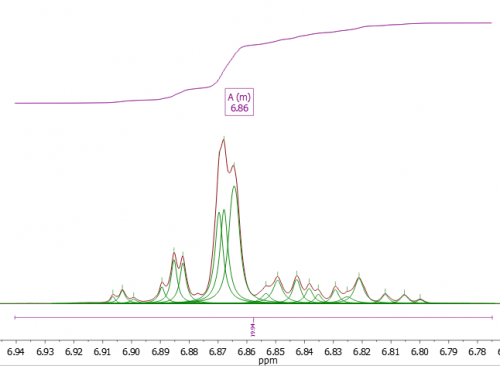
Fig. 2. A spectrum having quite high impurity levels. Here we compare results using the default impurity level threshold - 7.5% and an elevated one, 25%. For each we also compare standard automatic multiplet detection with Auto Assignments. For any given level of correctness Auto Assignments perform better than simple Auto Multiplets, and using a sensible threshold for this sample together with Auto Assignments (D) finally results in the correct multiplicities detected. In this case you should consider “Prioritise clean multiplets” as a filter.
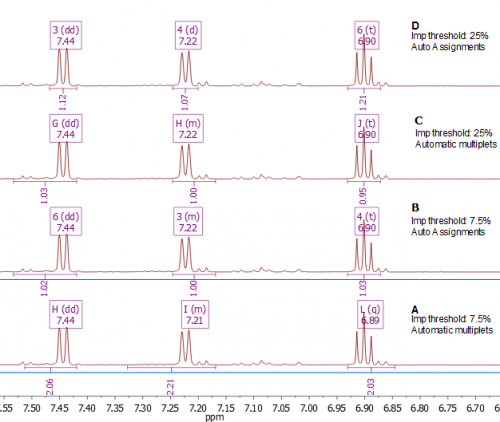
Fig. 3. In this spectrum the DMSO and residual water peaks were saturated. For the dataset it would be prudent to “Prioritise aromatic signals” as peaks in this region will not be partially saturated, and the baseline is less perturbed.

A detailed catalogue of the appropriate considerations for all data is impossible, but these examples hopefully give you a clue of what to look out for. Before implementing qNMR it is therefore a good idea to look at the effect of these selections and decide which are appropriate in your case. We can advise you if necessary.
Integral limits
It would be nice if the software automatically chose “sensible” integral limits. To this end, the user may specify a “multiplication factor”. The peak widths of the constituent multiplet are multiplied by this factor to derive a target integral width, or window. When multiplets are close to one another then naturally this must be truncated. This process further makes the entire process more robust under automation.
Mnova implementation
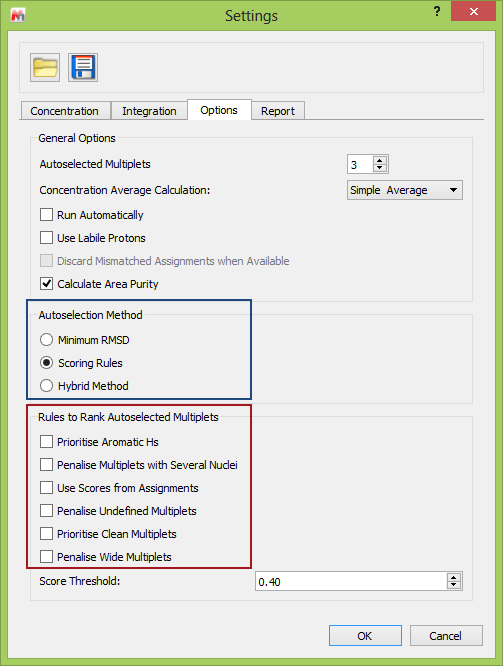
Using this functionality in Mnova is very easy, and the options are accessed through the qNMR “Settings” (see below). This shows (blue box) the overall method used to choose the multiplets, and in the red box the available multiplet penalty selections. Note than more than one Rule can be selected.
This exact same functionality is presented and applied when one might be analysing data manually, using the User Interface, automatically analysing a series of pre-acquired data (batch mode), or performing the analyses as the data are acquired by the spectrometer (listener mode).
Fig 4. The User Interface of Mnova showing the relevant options in the Settings.
Conclusions
We have shown that the quantification within Mnova offers useful alternatives to ensure that only the solute peaks are integrated, and that the “best” ones are chosen to determine the average concentration. Using this together with Auto Assignments makes for a powerful and robust workflow that is amenable to manual and automatic operation.