What's new in Mnova qNMR 2.0?
We have increased considerably the functionality of this module. A brief summary of the improvements is shown below:
Bug fixes to the basic quantitation module.
qNMR/Purity has seen significant enhancements:
- Create your custom experiments and store them in your own Libraries.
- Experiments can be applied to repeated analysis to ensure adhesion to your specified procedure.
- A new metadata reading capability of sample analysis-specific data, stored with the NMR data.
- Experiments can be used to analyze replicate spectra for a sample.
- The “Advised Analysis” functionality can do the analysis for you, making Purity determination a 1-click procedure.
qNMR overview
Modern qNMR is an exciting field already with a multitude of applications – and more to come. It is perhaps surprising that qNMR is reaching new audiences in the Analytical Chemistry community every day. This may be due to any number of a factors, including:
- Awareness
- Emergence of cheaper, easily sited, compact NMR systems
- Improved automatic spectrum analysis
The basic currency of qNMR rests of the very simple principle that applies to any high-resolution NMR spectrum: for any species, its concentration is proportional to the “normalized” integrals of any multiplet. A normalized integral is the integral divided by the number of nuclides from which it arises. qNMR can analyse (quite) pure, single compounds to determine the concentration or purity.
Highlights of the qNMR module:
- Mnova has been validated for qNMR use: Goedecke et al, Phytochem Anal (2013) 24, 581.
- Works on the basis of identified multiplets for a compound, and the number of Hs for each.
- A “response factor” is determined for a particular instrument and experimental setup, and is used without further reference.[Concentration]
- Automatic detection of all peaks to be excluded from the analysis: residual protic solvent, water, impurities.
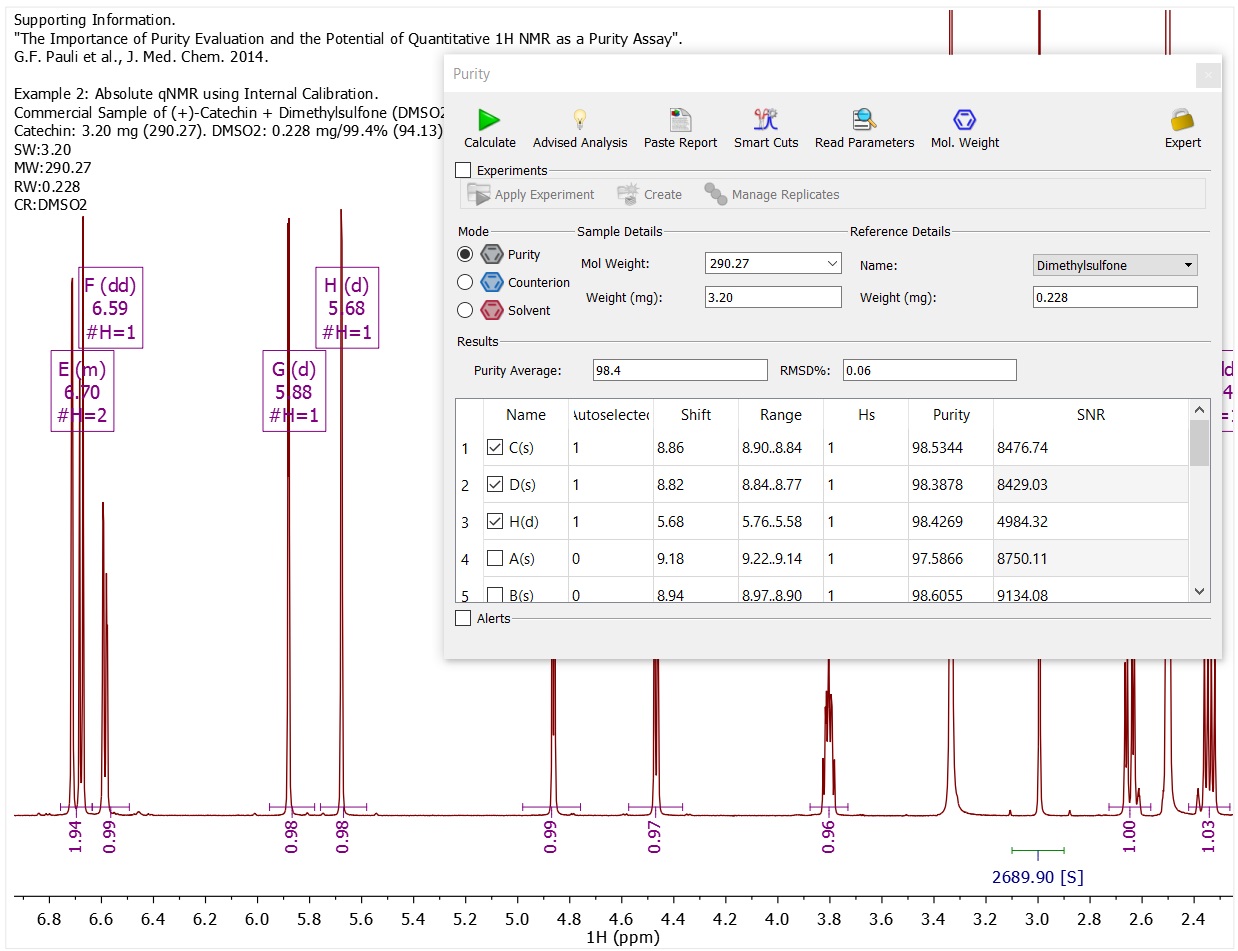
- For purity, input data for your own certified reference standard compounds.
- Save and reuse optimized analysis parameters.
- Easily manage replicate samples.[Purity]
- An optimized qNMR Purity analysis can be performed with a single button click.
- Save sample metadata (weights, etc.) with the NMR data, and read into the analysis.
- Define a Standard Operating Procedure (SOP) that can be used by others.